Bacterial Resistance to Antibiotics (page 1)
(This chapter has 4 pages)
Introduction

In the past 60 years, antibiotics have been critical in the fight against infectious disease caused by bacteria and other microbes. Antimicrobial chemotherapy has been a leading cause for the dramatic rise of average life expectancy in the Twentieth Century. However, disease-causing microbes that have become resistant to antibiotic drug therapy are an increasing public health problem. Wound infections, gonorrhea, tuberculosis, pneumonia, septicemia and childhood ear infections are just a few of the diseases that have become hard to treat with antibiotics. One part of the problem is that bacteria and other microbes that cause infections are remarkably resilient and have developed several ways to resist antibiotics and other antimicrobial drugs. Another part of the problem is due to increasing use, and misuse, of existing antibiotics in human and veterinary medicine and in agriculture.
In 1998, in the United States, 80 million prescriptions of antibiotics for human use were filled. This equals 12,500 tons in one year. Animal and agricultural uses of antibiotics are added to human use. Agricultural practices account for over 60% of antibiotic usage in the U.S., so this adds an additional 18,000 tons per year to the antibiotic burden in the environment.
Nowadays, about 70 percent of the bacteria that cause infections in hospitals are resistant to at least one of the drugs most commonly used for treatment. Some organisms are resistant to all approved antibiotics and can only be treated with experimental and potentially toxic drugs. An alarming increase in resistance of bacteria that cause community acquired infections has also been documented, especially in the staphylococci and pneumococci (Streptococcus pneumoniae), which are prevalent causes of disease and mortality. In a recent study, 25% of bacterial pneumonia cases were shown to be resistant to penicillin, and an additional 25% of cases were resistant to more than one antibiotic.
Microbial development of resistance, as well as economic incentives, has resulted in research and development in the search for new antibiotics in order to maintain a pool of effective drugs at all times. While the development of resistant strains is inevitable, the slack ways that we administer and use antibiotics has greatly exacerbated the process.
Unless antibiotic resistance problems are detected as they emerge, and actions are taken immediately to contain them, society could be faced with previously treatable diseases that have become again untreatable, as in the days before antibiotics were developed.
History of antibiotics and emergence of antibiotic resistance
The first antibiotic, penicillin, was discovered in 1929 by Sir Alexander Fleming, who observed inhibition of staphylococci on an agar plate contaminated by aPenicillium mold. Fleming was searching for potential antibacterial compounds. He noticed that a patch of the mold Penicillium notatum had grown on a plate containing the bacterium Staphylococcus and that around the mold there was a zone where no Staphylococcus could grow. After more research, he was able to show that culture broth of the mold prevented growth of the Staphylococcuseven when diluted up to 800 times. He named the active substance penicillin but was unable to isolate it.

In the center of the plate is a colony of Penicillium notatum, a mold that produces penicillin. After appearance of the mold colony, the plate was overlaid with a bacterial culture of Micrococcus luteus which forms a yellow "lawn" of growth. A zone of inhibition of bacterial growth surrounds the fungal colony where penicillin has diffused into the medium. http://helios.bto.ed.ac.uk/bto/microbes/penicill.htm#Top

While Fleming was working on penicillin, Gerhard Domagk, a German doctor, announced the discovery of a synthetic molecule with antibacterial properties. He named the compound Prontosil, and it became the first of a long series of synthetic antibiotics called sulfonamides or sulfa drugs. Prontosil was introduced to clinical use in the 1930s and was used to combat urinary tract infections, pneumonia and other conditions. While sulfa drugs in many cases are not as effective as natural antibiotics, they are now in widespread use for the treatment of many conditions. Gerhard Domagk was awarded the Nobel prize in 1939 for his discovery of Prontosil.In 1946, penicillin became generally available for treatment of bacterial infections, especially those caused by staphylococci and streptococci. Initially, the antibiotic was effective against all sorts of infections caused by these two Gram-positive bacteria. Penicillin had unbelievable ability to kill these bacterial pathogens without harming the host that harbored them. It is important to note that a significant fraction of all human infections are caused by these two bacteria (i.e., strep throat, pneumonia, scarlet fever, septicemia, skin infections, wound infections, etc.).
In the late 1940s and early 1950s, new antibiotics were introduced, including streptomycin, chloramphenicol and tetracycline, and the age of antibiotic chemotherapy came into full being. These antibiotics were effective against the full array of bacterial pathogens including Gram-positive and Gram-negative bacteria, intracellular parasites, and the tuberculosis bacillus. Synthetic antimicrobial agents such as the "sulfa drugs" (sulfonamides) and anti-tuberculosis drugs, such as para aminosalicylic acid (PAS) and isoniazid (INH), were also brought into wider usage.
The first signs of antibiotic resistance
There has probably been a gene pool in nature for resistance to antibiotic as long as there has been for antibiotic production, for most microbes that are antibiotic producers are resistant to their own antibiotic. In retrospect, it is not surprising that resistance to penicillin in some strains of staphylococci was recognized almost immediately after introduction of the drug in 1946. Likewise, very soon after their introduction in the late 1940s, resistance to streptomycin, chloramphenicol and tetracycline was noted. By 1953, during a Shigella outbreak in Japan, a strain of the dysentery bacillus (Shigella dysenteriae) was isolated which was multiple drug resistant, exhibiting resistance to chloramphenicol, tetracycline, streptomycin and the sulfonamides. Over the years, and continuing into the present almost every known bacterial pathogen has developed resistance to one or more antibiotics in clinical use.
Evidence also began to accumulate that bacteria could pass genes for drug resistance between strains and even between species. For example, antibiotic-resistance genes of staphylococci are carried on plasmids that can be exchanged with Bacillus, Streptococcus and Enterococcus providing the means for acquiring additional genes and gene combinations. Some are carried on transposons, segments of DNA that can exist either in the chromosome or in plasmids. In any case, it is clear that genes for antibiotic resistance can be exchanged between strains and species of bacteria by means of the processes of horizontal gene transmission (HGT).
Multiple drug resistant organisms
Multiple drug resistant organisms are resistant to treatment with several, often unrelated, antimicrobial agents as described above in Shigella. Some of the most important types of multiple drug resistant organisms that have been encountered include:
MRSA - methicillin/oxacillin-resistant Staphylococcus aureus
VRE - vancomycin-resistant enterococci
ESBLs - extended-spectrum beta-lactamases (which are resistant to cephalosporins and monobactams)
PRSP - penicillin-resistant Streptococcus pneumoniae
MRSA and VRE are the most commonly encountered multiple drug resistant organisms in patients residing in non-hospital healthcare facilities, such as nursing homes and other long-term care facilities. PRSP are more common in patients seeking care in outpatient settings such as physicians' offices and clinics, especially in pediatric settings. ESBLs are most often encountered in the hospital (intensive care) setting, but MRSA and VRE also have a significant nosocomial ecology.
Methicillin-Resistant Staph Aureus. MRSA refers to "methicillin-resistantStaphylococcus aureus", which are strains of the bacterium that are resistant to the action of methicillin, and related beta-lactam antibiotics (e.g. penicillin and cephalosporin). MRSA have evolved resistance not only to beta-lactam antibiotics, but to several classes of antibiotics. Some MRSA are resistant to all but one or two antibiotics, notably vancomycin-resistant. But there have been several reports of VRSA (Vancomycin-Resistant Staph Aureus) that are troublesome in the ongoing battle against staph infections.
MRSA are often sub-categorized as Hospital-Associated MRSA (HA-MRSA) or Community-Associated MRSA (CA-MRSA), depending upon the circumstances of acquiring disease. Based on current data, these are distinct strains of the bacterial species.
HA-MRSA occurs most frequently among patients who undergo invasive medical procedures or who have weakened immune systems and are being treated in hospitals and healthcare facilities such as nursing homes and dialysis centers. MRSA in healthcare settings commonly causes serious and potentially life threatening infections, such as bloodstream infections, surgical site infections or pneumonia.
In the case of HA- MRSA, patients who already have an MRSA infection or who carry the bacteria on their bodies but do not have symptoms (colonized) are the most common sources of transmission. The main mode of transmission to other patients is through human hands, especially healthcare workers' hands. Hands may become contaminated with MRSA bacteria by contact with infected or colonized patients. If appropriate hand hygiene such as washing with soap and water or using an alcohol-based hand sanitizer is not performed, the bacteria can be spread when the healthcare worker touches other patients.
MRSA infections that occur in otherwise healthy people who have not been recently (within the past year) hospitalized or had a medical procedure (such as dialysis, surgery, catheters) are categorized as community-associated (CA-MRSA) infections. These infections are usually skin infections, such as abscesses, boils, and other pus-filled lesions.
About 75 percent of CA-MRSA infections are localized to skin and soft tissue and usually can be treated effectively. However, CA-MRSA strains display enhanced virulence, spread more rapidly and cause more severe illness than traditional HA-MRSA infections, and can affect vital organs leading to widespread infection (sepsis), toxic shock syndrome and pneumonia. It is not known why some healthy people develop CA-MRSA skin infections that are treatable whereas others infected with the same strain develop severe, fatal infections.
Studies have shown that rates of CA-MRSA infection are growing fast. One study of children in south Texas found that cases of CA-MRSA had a 14-fold increase between 1999 and 2001.
CA-MRSA skin infections have been identified among certain populations that share close quarters or experience more skin-to-skin contact. Examples are team athletes, military recruits, and prisoners. However, more and more CA-MRSA infections are being seen in the general community as well, especially in certain geographic regions.
Also, CA-MRSA are infecting much younger people. In a study of Minnesotans published in The Journal of the American Medical Association, the average age of people with MRSA in a hospital or healthcare facility was 68. But the average age of a person with CA-MRSA was only 23.
More people in the U.S. now die from MRSA infection than from AIDS. Methicillin-resistant Staphylococcus aureus was responsible for an estimated 94,000 life-threatening infections and 18,650 deaths in 2005, as reported by CDC in the Oct. 17, 2007 issue of The Journal of the American Medical Association. The national estimate is more than double the invasive MRSA prevalence reported five years earlier. That same year, roughly 16,000 people in the U.S. died from AIDS, according to CDC. While most invasive MRSA infections could be traced to a hospital stay or some other health care exposure, about 15% of invasive infections occurred in people with no known health care risk. Two-thirds of the 85% of MRSA infections that could be traced to hospital stays or other health care exposures occurred among people who were no longer hospitalized. People over age 65 were four times more likely than the general population to get an MRSA infection. Incidence rates among blacks were twice that of the general population, and rates were lowest among children over the age of 4 and teens.
ESBL producing strains have been isolated from abscesses, blood, catheter tips, lung, peritoneal fluid, sputum, and throat cultures. They apparently have a world-wide distribution. Rates of isolation vary greatly worldwide and within geographic areas and are rapidly changing over time. In the United States, between 1990 to 1993, a survey of the intensive care units of 400 hospitals recorded an increase from 3.6% to 14.4% in ESBL producing strains ofKlebsiella. In 1994, the CDC reported that 8% of Klebsiella spp from a few large centers produced ESBLs. In Europe, as of 1995, ESBLs occurred in 20%-25% ofKlebsiella ssp from patients in ICUs, although they were found in patients up to 30%-40% frequency in France.
Known risk factors for colonization and/or infection with organisms harboring ESBLs include admission to an intensive care unit, recent surgery, instrumentation, prolonged hospital stay and antibiotic exposure, especially to extended-spectrum beta-lactam antibiotics. Use of extended-spectrum antibiotics exerts a selective pressure for emergence of ESBL producing strains. The resistance plasmids can then be transferred to other bacteria, not necessarily of the same species, conferring resistance to them.
The lower GI tract of colonized patients is the main reservoir of these organisms. Gastrointestinal carriage can persist for months. In some cities in the United States, nursing homes may be an important reservoir of ESBL producing strains. Nursing home patients are more likely to be treated empirically with antibiotics, and thus on admission to a hospital to be more likely to possess an ESBL producing strain. Patient to patient transmission of ESBL producing organisms occurs via the hands of hospital staff. It is known that ESBL producing strains can survive in the hospital environment.
Nosocomial infections in patients occur through the administration of extended spectrum beta-lactam antibiotics or via transmission from other patients via health care workers, who become colonized with resistant strains via exposure to patients or other health care workers. Spread of ESBL producing strains can be minimized by good infection control practices, especially by good hand washing technique.
Bacterial mechanisms of antibiotic resistance
Several mechanisms have evolved in bacteria which confer them with antibiotic resistance. These mechanisms can either chemically modify the antibiotic, render it inactive through physical removal from the cell, or modify target site so that it is not recognized by the antibiotic.
The most common mode is enzymatic inactivation of the antibiotic. An existing cellular enzyme is modified to react with the antibiotic in such a way that it no longer affects the microorganism. An alternative strategy utilized by many bacteria is the alteration of the antibiotic target site. These and other mechanisms are shown in the the figure and accompanying table below.
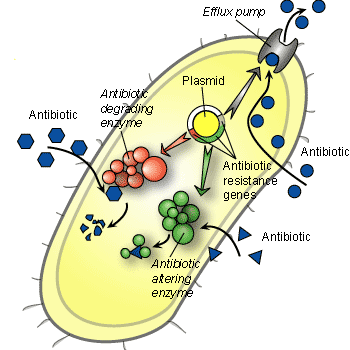
Three mechanisms of antibiotic resistance in bacteria. Most, but not all, resistance mechanisms are encoded by plasmids, which are potentially transmissible to other bacteria. Clockwise. 12 o'clock: Efflux pumps are high-affinity reverse transport systems located in the membrane that transport the antibiotic out of the cell. This is the mechanism of resistance to tetracycline. 4 o'clock: A specific enzyme modifies the antibiotic in a way that it loses its activity. In the case of streptomycin, the antibiotic is chemically modified so that it will no longer bind to the ribosome to block protein synthesis. 9 o'clock: An enzyme is produced that degrades the antibiotic, thereby inactivating it. For example, the penicillinases are a group of beta-lactamase enzymes that cleave the beta lactam ring of the penicillin molecule.
The acquisition and spread of antibiotic resistance in bacteria
The development of resistance is inevitable following the introduction of a new antibiotic. Initial rates of resistance to new drugs are normally on the order of 1%. However, modern uses of antibiotics have caused a huge increase in the number of resistant bacteria. In fact, within 8-12 years after wide-spread use, strains resistant to multiple drugs become widespread. Multiple drug resistant strains of some bacteria have reached the proportion that virtually no antibiotics are available for treatment.
Antibiotic resistance in bacteria may be an inherent trait of the organism (e.g. a particular type of cell wall structure) that renders it naturally resistant, or it may be acquired by means of mutation in its own DNA or acquisition of resistance-conferring DNA from another source.
Inherent (natural) resistance. Bacteria may be inherently resistant to an antibiotic. For example, an organism lacks a transport system for an antibiotic; or an organism lacks the target of the antibiotic molecule; or, as in the case of Gram-negative bacteria, the cell wall is covered with an outer membrane that establishes a permeability barrier against the antibiotic.
Acquired resistance. Several mechanisms are developed by bacteria in order to acquire resistance to antibiotics. All require either the modification of existing genetic material or the acquisition of new genetic material from another source.
Vertical gene transfer
The spontaneous mutation frequency for antibiotic resistance is on the order of about of about 10-8- 10-9. This means that one in every every 108- 109 bacteria in an infection will develop resistance through the process of mutation. In E. coli, it has been estimated that streptomycin resistance is acquired at a rate of approximately 10-9 when exposed to high concentrations of streptomycin. Although mutation is a very rare event, the very fast growth rate of bacteria and the absolute number of cells attained means that it doesn't take long before resistance is developed in a population.
Once the resistance genes have developed, they are transferred directly to all the bacteria's progeny during DNA replication. This is known as vertical gene transfer or vertical evolution. The process is strictly a matter of Darwinian evolution driven by principles of natural selection: a spontaneous mutation in the bacterial chromosome imparts resistance to a member of the bacterial population. In the selective environment of the antibiotic, the wild type (non mutants) are killed and the resistant mutant is allowed to grow and flourish
Horizontal gene transfer
Another mechanism beyond spontaneous mutation is responsible for the acquisition of antibiotic resistance. Lateral or horizontal gene transfer (HGT) is a process whereby genetic material contained in small packets of DNA can be transferred between individual bacteria of the same species or even between different species.

Mechanisms of horizontal gene transfer (HGT) in bacteria
The combined effects of fast growth rates to large densities of cells, genetic processes of mutation and selection, and the ability to exchange genes, account for the extraordinary rates of adaptation and evolution that can be observed in the bacteria. For these reasons bacterial adaptation (resistance) to the antibiotic environment seems to take place very rapidly in evolutionary time. Bacteria evolve fast!

Tests for sensitivity and resistance to antibiotics. (Left) The size of the zones of inhibition of microbial growth surrounding the antibiotic disks on the plate are an indication of microbial susceptibility to the antibiotic. (Right) By the use of these disks it is also possible to detect the occurrence of individual mutants within the culture that have developed antibiotic resistance. This image shows a close-up of the novobiocin disk (marked by an arrow on the whole plate) near which individual mutant cells in the bacterial population that were resistant to the antibiotic and have given rise to small colonies within the zone of inhibition.
Societal, medical and agricultural practices that lead to antibiotic resistance
In the face of a microbe's inherent ability to develop antibiotic resistance, many societal. medical and agricultural practices contribute to this process, foremost of which are discussed below.
Antibiotics in food and water
Prescription drugs are not the only source of antibiotics in the environment. In the United States, antibiotics can be found in beef cattle, pigs and poultry. The same antibiotics then find their way into municipal water systems when the runoff from housing facilities and feedlots contaminates streams and groundwater. So it's a double hit: we get antibiotics in our food and drinking water, and we meanwhile promote bacterial resistance. Routine feeding of antibiotics to animals is banned in the European Union and many other industrialized countries. Maybe they know something we don't.
Indiscriminate use of antibiotics in agriculture and veterinary practice
The non-therapeutic use of antibiotics in livestock production makes up at least 60 percent of the total antimicrobial production in the United States. Irresponsible use of antibiotics in farm animals can lead to the development of resistance in bacteria associated with the animal or with people who eat the animal. Such resistance can then be passed on to human pathogens by mechanisms of HGT.
Of major concern is the use of antibiotics as feed additives given to farm animals to promote animal growth and to prevent infections (rather than cure infections). The use of an antibiotic in this way contributes to the emergence of antibiotic-resistant pathogens and reduces the effectiveness of the antibiotic to combat human infections.
Antibiotic resistance in genetically modified crops
Antibiotic-resistance genes are used as "markers" in genetically modified crops. The genes are inserted into the plant in early stages of development to in order to detect specific genes of interest . e.g. herbicide-resistant genes or insecticidal toxin genes. The antibiotic-resistance genes have no further role to play, but they are not removed from the final product. This practice has met with criticism because of the potential that the antibiotic-resistance genes could be acquired by microbes in the environment. In some cases these marker genes confer resistance to front-line antibiotics such as the beta-lactams and aminoglycosides.
Inappropriate use of antibiotics in the medical environment
One problem is the casual use of antibiotics in medical situations where they are of no value. This is the fault of both health care workers and patients. Prescribers sometimes thoughtlessly prescribe 'informed' demanding patients with antibiotics. This leads to use of antibiotics in circumstances where they are of not needed, e.g. viral upper respiratory infections such as cold and flu, except when there is serious threat of secondary bacterial infection. Another problem is patient failure to adhere to regimens for prescribed antibiotics.
Patients and doctors need to realize their responsibility when they begin an antibiotic regimen to combat an infectious disease. There are several measures that should be considered.
� Patients should not take antibiotics for which there is no medical value (corollary: doctors should not prescribe antibiotics for which there is no medical value).
� Patients should adhere to appropriate prescribing guidelines and take antibiotics until they have finished.
� Patients should be give combinations of antibiotics, when necessary, to minimize the development of resistance to a single antibiotic (as in the case of TB).
� Patients need to be given another antibiotic or combination of antibiotics if the first is not working.
Combating antibiotic resistance
The following are recommendations to combat the development of antibiotic resistance in bacteria and other microorganisms.
Search for new antibiotics. To combat the occurrence of resistant bacteria, biotechnology and pharmaceutical companies must constantly research, develop and test new antimicrobials in order to maintain a pool of effective drugs on the market.
Stop the use of antibiotics as growth-promoting substances in farm animals. Of major concern is the use of antibiotics as feed additives given to farm animals to promote animal growth and to prevent infections rather than cure infections. The use of such antibiotics contributes to the emergence of antibiotic-resistant bacteria that threaten human health and decreases the effectiveness of the same antibiotics used to combat human infections.
Use the right antibiotic in an infectious situation as determined by antibiotic sensitivity testing, when possible.
Large scale public health education efforts are underway to stress the importance of finishing prescriptions. Indeed, in many places, failure to finish tuberculosis prescriptions can result in jail time.
Summary
The discovery of antibiotics was a leap in modern medicine. They have been able to stop the growth or kill many different kinds of microorganisms. However, bacteria have proven to be much more innovative and adaptive than we imagined and have developed resistance to antibiotics at an ever increasing pace. Bad practices and mismanagement have only exacerbated the situation. We could soon return to a state of medical health that was as dire as that which occurred prior to antibiotic use. However, with more research, education of the public, and well thought out regulations, the problems
can be solved. Several strategies are currently used to find new antibacterial compounds and new strategies are in development and trial.
Not only is there a problem in finding new antibiotics to fight old diseases (because resistant strains of bacteria have emerged), there is a parallel problem to find new antibiotics to fight new diseases. In the past three decades, many "new" bacterial diseases have been discovered (E. coli O157:H7 gastric ulcers, Lyme disease, toxic shock syndrome, "skin-eating" streptococci). Already broad patterns of resistance exist in these pathogens, and it seems likely that we will soon need new antibiotics to replace the handful that are effective now against these bacteria, especially as resistance begins to emerge among them in the selective environment antibiotic chemotherapy.
It is said that the discovery and use of antibiotics and immunization procedures against infectious disease are two developments in the field of microbiology that have contributed about twenty years to the average life span of humans in developed countries where these practices are employed. While the greater part of this span in time is probably due to vaccination, most of us are either still alive or have family members or friends who are still alive because an antibiotic conquered an infection that otherwise would have killed them. If we want to retain this medical luxury in our society we must be vigilant and proactive We must fully understand how and why antimicrobial agents work, and why they don't work, and realize that we must maintain a stride ahead of microbial pathogens that can only be contained by antibiotic chemotherapy.
END OF CHAPTER
(This chapter has 4 pages)
Introduction

In the past 60 years, antibiotics have been critical in the fight against infectious disease caused by bacteria and other microbes. Antimicrobial chemotherapy has been a leading cause for the dramatic rise of average life expectancy in the Twentieth Century. However, disease-causing microbes that have become resistant to antibiotic drug therapy are an increasing public health problem. Wound infections, gonorrhea, tuberculosis, pneumonia, septicemia and childhood ear infections are just a few of the diseases that have become hard to treat with antibiotics. One part of the problem is that bacteria and other microbes that cause infections are remarkably resilient and have developed several ways to resist antibiotics and other antimicrobial drugs. Another part of the problem is due to increasing use, and misuse, of existing antibiotics in human and veterinary medicine and in agriculture.
In 1998, in the United States, 80 million prescriptions of antibiotics for human use were filled. This equals 12,500 tons in one year. Animal and agricultural uses of antibiotics are added to human use. Agricultural practices account for over 60% of antibiotic usage in the U.S., so this adds an additional 18,000 tons per year to the antibiotic burden in the environment.
Nowadays, about 70 percent of the bacteria that cause infections in hospitals are resistant to at least one of the drugs most commonly used for treatment. Some organisms are resistant to all approved antibiotics and can only be treated with experimental and potentially toxic drugs. An alarming increase in resistance of bacteria that cause community acquired infections has also been documented, especially in the staphylococci and pneumococci (Streptococcus pneumoniae), which are prevalent causes of disease and mortality. In a recent study, 25% of bacterial pneumonia cases were shown to be resistant to penicillin, and an additional 25% of cases were resistant to more than one antibiotic.
Microbial development of resistance, as well as economic incentives, has resulted in research and development in the search for new antibiotics in order to maintain a pool of effective drugs at all times. While the development of resistant strains is inevitable, the slack ways that we administer and use antibiotics has greatly exacerbated the process.
Unless antibiotic resistance problems are detected as they emerge, and actions are taken immediately to contain them, society could be faced with previously treatable diseases that have become again untreatable, as in the days before antibiotics were developed.
History of antibiotics and emergence of antibiotic resistance
The first antibiotic, penicillin, was discovered in 1929 by Sir Alexander Fleming, who observed inhibition of staphylococci on an agar plate contaminated by aPenicillium mold. Fleming was searching for potential antibacterial compounds. He noticed that a patch of the mold Penicillium notatum had grown on a plate containing the bacterium Staphylococcus and that around the mold there was a zone where no Staphylococcus could grow. After more research, he was able to show that culture broth of the mold prevented growth of the Staphylococcuseven when diluted up to 800 times. He named the active substance penicillin but was unable to isolate it.

In the center of the plate is a colony of Penicillium notatum, a mold that produces penicillin. After appearance of the mold colony, the plate was overlaid with a bacterial culture of Micrococcus luteus which forms a yellow "lawn" of growth. A zone of inhibition of bacterial growth surrounds the fungal colony where penicillin has diffused into the medium. http://helios.bto.ed.ac.uk/bto/microbes/penicill.htm#Top
Several years later, in 1939, Ernst Chain and Howard Florey developed a way to isolate penicillin and used it to treat bacterial infections during the Second World War. The new drug came into clinical usage in 1946 and made a huge impact on public health. For these discoveries Fleming, Chain and Florey were awarded the Nobel prize in 1945. Their discovery and development revolutionized modern medicine and paved the way for the development of many more natural antibiotics.

While Fleming was working on penicillin, Gerhard Domagk, a German doctor, announced the discovery of a synthetic molecule with antibacterial properties. He named the compound Prontosil, and it became the first of a long series of synthetic antibiotics called sulfonamides or sulfa drugs. Prontosil was introduced to clinical use in the 1930s and was used to combat urinary tract infections, pneumonia and other conditions. While sulfa drugs in many cases are not as effective as natural antibiotics, they are now in widespread use for the treatment of many conditions. Gerhard Domagk was awarded the Nobel prize in 1939 for his discovery of Prontosil.
In the late 1940s and early 1950s, new antibiotics were introduced, including streptomycin, chloramphenicol and tetracycline, and the age of antibiotic chemotherapy came into full being. These antibiotics were effective against the full array of bacterial pathogens including Gram-positive and Gram-negative bacteria, intracellular parasites, and the tuberculosis bacillus. Synthetic antimicrobial agents such as the "sulfa drugs" (sulfonamides) and anti-tuberculosis drugs, such as para aminosalicylic acid (PAS) and isoniazid (INH), were also brought into wider usage.
The first signs of antibiotic resistance
There has probably been a gene pool in nature for resistance to antibiotic as long as there has been for antibiotic production, for most microbes that are antibiotic producers are resistant to their own antibiotic. In retrospect, it is not surprising that resistance to penicillin in some strains of staphylococci was recognized almost immediately after introduction of the drug in 1946. Likewise, very soon after their introduction in the late 1940s, resistance to streptomycin, chloramphenicol and tetracycline was noted. By 1953, during a Shigella outbreak in Japan, a strain of the dysentery bacillus (Shigella dysenteriae) was isolated which was multiple drug resistant, exhibiting resistance to chloramphenicol, tetracycline, streptomycin and the sulfonamides. Over the years, and continuing into the present almost every known bacterial pathogen has developed resistance to one or more antibiotics in clinical use.
Evidence also began to accumulate that bacteria could pass genes for drug resistance between strains and even between species. For example, antibiotic-resistance genes of staphylococci are carried on plasmids that can be exchanged with Bacillus, Streptococcus and Enterococcus providing the means for acquiring additional genes and gene combinations. Some are carried on transposons, segments of DNA that can exist either in the chromosome or in plasmids. In any case, it is clear that genes for antibiotic resistance can be exchanged between strains and species of bacteria by means of the processes of horizontal gene transmission (HGT).
Multiple drug resistant organisms
Multiple drug resistant organisms are resistant to treatment with several, often unrelated, antimicrobial agents as described above in Shigella. Some of the most important types of multiple drug resistant organisms that have been encountered include:
MRSA - methicillin/oxacillin-resistant Staphylococcus aureus
VRE - vancomycin-resistant enterococci
ESBLs - extended-spectrum beta-lactamases (which are resistant to cephalosporins and monobactams)
PRSP - penicillin-resistant Streptococcus pneumoniae
MRSA and VRE are the most commonly encountered multiple drug resistant organisms in patients residing in non-hospital healthcare facilities, such as nursing homes and other long-term care facilities. PRSP are more common in patients seeking care in outpatient settings such as physicians' offices and clinics, especially in pediatric settings. ESBLs are most often encountered in the hospital (intensive care) setting, but MRSA and VRE also have a significant nosocomial ecology.
Methicillin-Resistant Staph Aureus. MRSA refers to "methicillin-resistantStaphylococcus aureus", which are strains of the bacterium that are resistant to the action of methicillin, and related beta-lactam antibiotics (e.g. penicillin and cephalosporin). MRSA have evolved resistance not only to beta-lactam antibiotics, but to several classes of antibiotics. Some MRSA are resistant to all but one or two antibiotics, notably vancomycin-resistant. But there have been several reports of VRSA (Vancomycin-Resistant Staph Aureus) that are troublesome in the ongoing battle against staph infections.
MRSA are often sub-categorized as Hospital-Associated MRSA (HA-MRSA) or Community-Associated MRSA (CA-MRSA), depending upon the circumstances of acquiring disease. Based on current data, these are distinct strains of the bacterial species.
HA-MRSA occurs most frequently among patients who undergo invasive medical procedures or who have weakened immune systems and are being treated in hospitals and healthcare facilities such as nursing homes and dialysis centers. MRSA in healthcare settings commonly causes serious and potentially life threatening infections, such as bloodstream infections, surgical site infections or pneumonia.
In the case of HA- MRSA, patients who already have an MRSA infection or who carry the bacteria on their bodies but do not have symptoms (colonized) are the most common sources of transmission. The main mode of transmission to other patients is through human hands, especially healthcare workers' hands. Hands may become contaminated with MRSA bacteria by contact with infected or colonized patients. If appropriate hand hygiene such as washing with soap and water or using an alcohol-based hand sanitizer is not performed, the bacteria can be spread when the healthcare worker touches other patients.
MRSA infections that occur in otherwise healthy people who have not been recently (within the past year) hospitalized or had a medical procedure (such as dialysis, surgery, catheters) are categorized as community-associated (CA-MRSA) infections. These infections are usually skin infections, such as abscesses, boils, and other pus-filled lesions.
About 75 percent of CA-MRSA infections are localized to skin and soft tissue and usually can be treated effectively. However, CA-MRSA strains display enhanced virulence, spread more rapidly and cause more severe illness than traditional HA-MRSA infections, and can affect vital organs leading to widespread infection (sepsis), toxic shock syndrome and pneumonia. It is not known why some healthy people develop CA-MRSA skin infections that are treatable whereas others infected with the same strain develop severe, fatal infections.
Studies have shown that rates of CA-MRSA infection are growing fast. One study of children in south Texas found that cases of CA-MRSA had a 14-fold increase between 1999 and 2001.
CA-MRSA skin infections have been identified among certain populations that share close quarters or experience more skin-to-skin contact. Examples are team athletes, military recruits, and prisoners. However, more and more CA-MRSA infections are being seen in the general community as well, especially in certain geographic regions.
Also, CA-MRSA are infecting much younger people. In a study of Minnesotans published in The Journal of the American Medical Association, the average age of people with MRSA in a hospital or healthcare facility was 68. But the average age of a person with CA-MRSA was only 23.
More people in the U.S. now die from MRSA infection than from AIDS. Methicillin-resistant Staphylococcus aureus was responsible for an estimated 94,000 life-threatening infections and 18,650 deaths in 2005, as reported by CDC in the Oct. 17, 2007 issue of The Journal of the American Medical Association. The national estimate is more than double the invasive MRSA prevalence reported five years earlier. That same year, roughly 16,000 people in the U.S. died from AIDS, according to CDC. While most invasive MRSA infections could be traced to a hospital stay or some other health care exposure, about 15% of invasive infections occurred in people with no known health care risk. Two-thirds of the 85% of MRSA infections that could be traced to hospital stays or other health care exposures occurred among people who were no longer hospitalized. People over age 65 were four times more likely than the general population to get an MRSA infection. Incidence rates among blacks were twice that of the general population, and rates were lowest among children over the age of 4 and teens.
Extended-Spectrum beta-lactamase (ESBL) - producing Gram-negative bacteria Extended-spectrum beta-lactamases (ESBLs) are plasmid-associated beta lactamases that have recently been found in the Enterobacteriaceae. ESBLs are capable of hydrolyzing penicillins, many narrow spectrum cephalosporins, many extended-spectrum cephalosporins, oxyimino-cephalosporins (cefotaxime, ceftazidime), and monobactams (aztreonam). Beta-lactamase inhibitors (e.g. clavulanic acid) generally inhibit ESBL producing strains. ESBL producing isolates are most commonly Klebsiella ssp, predominantly Klebsiella pneumoniae, and E. coli, but they have been found throughout the Enterobacteriaeae.
Because ESBL enzymes are plasmid mediated, the genes encoding these enzymes are easily transferable among different bacteria. Most of these plasmids not only contain DNA encoding ESBL enzymes but also carry genes conferring resistance to several non-ß-Lactam antibiotics. Consequently, most ESBL isolates are resistant to many classes of antibiotics. The most frequent coresistances found in ESBL-producing organisms are aminoglycosides, fluoroquinolones, tetracyclines, chloramphenicol, and sulfamethoxazole-trimethoprim. Treatment of these multiple drug-resistant organisms is a therapeutic challenge.ESBL producing strains have been isolated from abscesses, blood, catheter tips, lung, peritoneal fluid, sputum, and throat cultures. They apparently have a world-wide distribution. Rates of isolation vary greatly worldwide and within geographic areas and are rapidly changing over time. In the United States, between 1990 to 1993, a survey of the intensive care units of 400 hospitals recorded an increase from 3.6% to 14.4% in ESBL producing strains ofKlebsiella. In 1994, the CDC reported that 8% of Klebsiella spp from a few large centers produced ESBLs. In Europe, as of 1995, ESBLs occurred in 20%-25% ofKlebsiella ssp from patients in ICUs, although they were found in patients up to 30%-40% frequency in France.
Known risk factors for colonization and/or infection with organisms harboring ESBLs include admission to an intensive care unit, recent surgery, instrumentation, prolonged hospital stay and antibiotic exposure, especially to extended-spectrum beta-lactam antibiotics. Use of extended-spectrum antibiotics exerts a selective pressure for emergence of ESBL producing strains. The resistance plasmids can then be transferred to other bacteria, not necessarily of the same species, conferring resistance to them.
The lower GI tract of colonized patients is the main reservoir of these organisms. Gastrointestinal carriage can persist for months. In some cities in the United States, nursing homes may be an important reservoir of ESBL producing strains. Nursing home patients are more likely to be treated empirically with antibiotics, and thus on admission to a hospital to be more likely to possess an ESBL producing strain. Patient to patient transmission of ESBL producing organisms occurs via the hands of hospital staff. It is known that ESBL producing strains can survive in the hospital environment.
Nosocomial infections in patients occur through the administration of extended spectrum beta-lactam antibiotics or via transmission from other patients via health care workers, who become colonized with resistant strains via exposure to patients or other health care workers. Spread of ESBL producing strains can be minimized by good infection control practices, especially by good hand washing technique.
Bacterial mechanisms of antibiotic resistance
Several mechanisms have evolved in bacteria which confer them with antibiotic resistance. These mechanisms can either chemically modify the antibiotic, render it inactive through physical removal from the cell, or modify target site so that it is not recognized by the antibiotic.
The most common mode is enzymatic inactivation of the antibiotic. An existing cellular enzyme is modified to react with the antibiotic in such a way that it no longer affects the microorganism. An alternative strategy utilized by many bacteria is the alteration of the antibiotic target site. These and other mechanisms are shown in the the figure and accompanying table below.

Three mechanisms of antibiotic resistance in bacteria. Most, but not all, resistance mechanisms are encoded by plasmids, which are potentially transmissible to other bacteria. Clockwise. 12 o'clock: Efflux pumps are high-affinity reverse transport systems located in the membrane that transport the antibiotic out of the cell. This is the mechanism of resistance to tetracycline. 4 o'clock: A specific enzyme modifies the antibiotic in a way that it loses its activity. In the case of streptomycin, the antibiotic is chemically modified so that it will no longer bind to the ribosome to block protein synthesis. 9 o'clock: An enzyme is produced that degrades the antibiotic, thereby inactivating it. For example, the penicillinases are a group of beta-lactamase enzymes that cleave the beta lactam ring of the penicillin molecule.
| Antibiotic | Method of resistance | ||
| Chloramphenicol | reduced uptake into cell | ||
| Tetracycline | active efflux from the cell | ||
| β-lactams, Erythromycin, Lincomycin | eliminates or reduces binding of antibiotic to cell target | ||
| β-lactams, Aminoglycosides, Chloramphenicol | enzymatic cleavage or modification to inactivate antibiotic molecule | ||
| Sulfonamides, Trimethoprim | metabolic bypass of inhibited reaction | ||
| Sulfonamides, Trimethoprim | overproduction of antibiotic target (titration) | ||
The acquisition and spread of antibiotic resistance in bacteria
The development of resistance is inevitable following the introduction of a new antibiotic. Initial rates of resistance to new drugs are normally on the order of 1%. However, modern uses of antibiotics have caused a huge increase in the number of resistant bacteria. In fact, within 8-12 years after wide-spread use, strains resistant to multiple drugs become widespread. Multiple drug resistant strains of some bacteria have reached the proportion that virtually no antibiotics are available for treatment.
Antibiotic resistance in bacteria may be an inherent trait of the organism (e.g. a particular type of cell wall structure) that renders it naturally resistant, or it may be acquired by means of mutation in its own DNA or acquisition of resistance-conferring DNA from another source.
Inherent (natural) resistance. Bacteria may be inherently resistant to an antibiotic. For example, an organism lacks a transport system for an antibiotic; or an organism lacks the target of the antibiotic molecule; or, as in the case of Gram-negative bacteria, the cell wall is covered with an outer membrane that establishes a permeability barrier against the antibiotic.
Acquired resistance. Several mechanisms are developed by bacteria in order to acquire resistance to antibiotics. All require either the modification of existing genetic material or the acquisition of new genetic material from another source.
Vertical gene transfer
The spontaneous mutation frequency for antibiotic resistance is on the order of about of about 10-8- 10-9. This means that one in every every 108- 109 bacteria in an infection will develop resistance through the process of mutation. In E. coli, it has been estimated that streptomycin resistance is acquired at a rate of approximately 10-9 when exposed to high concentrations of streptomycin. Although mutation is a very rare event, the very fast growth rate of bacteria and the absolute number of cells attained means that it doesn't take long before resistance is developed in a population.
Once the resistance genes have developed, they are transferred directly to all the bacteria's progeny during DNA replication. This is known as vertical gene transfer or vertical evolution. The process is strictly a matter of Darwinian evolution driven by principles of natural selection: a spontaneous mutation in the bacterial chromosome imparts resistance to a member of the bacterial population. In the selective environment of the antibiotic, the wild type (non mutants) are killed and the resistant mutant is allowed to grow and flourish
Horizontal gene transfer
Another mechanism beyond spontaneous mutation is responsible for the acquisition of antibiotic resistance. Lateral or horizontal gene transfer (HGT) is a process whereby genetic material contained in small packets of DNA can be transferred between individual bacteria of the same species or even between different species.
There are at least three possible mechanisms of HGT, equivalent to the three processes of genetic exchange in bacteria. These are transduction, transformation or conjugation.
Conjugation occurs when there is direct cell-cell contact between two bacteria (which need not be closely related) and transfer of small pieces of DNA called plasmids takes place. This is thought to be the main mechanism of HGT.Transformation is a process where parts of DNA are taken up by the bacteria from the external environment. This DNA is normally present in the external environment due to the death and lysis of another bacterium.
Transduction occurs when bacteria-specific viruses (bacteriophages) transfer DNA between two closely related bacteria.

Mechanisms of horizontal gene transfer (HGT) in bacteria
The combined effects of fast growth rates to large densities of cells, genetic processes of mutation and selection, and the ability to exchange genes, account for the extraordinary rates of adaptation and evolution that can be observed in the bacteria. For these reasons bacterial adaptation (resistance) to the antibiotic environment seems to take place very rapidly in evolutionary time. Bacteria evolve fast!

Tests for sensitivity and resistance to antibiotics. (Left) The size of the zones of inhibition of microbial growth surrounding the antibiotic disks on the plate are an indication of microbial susceptibility to the antibiotic. (Right) By the use of these disks it is also possible to detect the occurrence of individual mutants within the culture that have developed antibiotic resistance. This image shows a close-up of the novobiocin disk (marked by an arrow on the whole plate) near which individual mutant cells in the bacterial population that were resistant to the antibiotic and have given rise to small colonies within the zone of inhibition.
Societal, medical and agricultural practices that lead to antibiotic resistance
In the face of a microbe's inherent ability to develop antibiotic resistance, many societal. medical and agricultural practices contribute to this process, foremost of which are discussed below.
Antibiotics in food and water
Prescription drugs are not the only source of antibiotics in the environment. In the United States, antibiotics can be found in beef cattle, pigs and poultry. The same antibiotics then find their way into municipal water systems when the runoff from housing facilities and feedlots contaminates streams and groundwater. So it's a double hit: we get antibiotics in our food and drinking water, and we meanwhile promote bacterial resistance. Routine feeding of antibiotics to animals is banned in the European Union and many other industrialized countries. Maybe they know something we don't.
Indiscriminate use of antibiotics in agriculture and veterinary practice
The non-therapeutic use of antibiotics in livestock production makes up at least 60 percent of the total antimicrobial production in the United States. Irresponsible use of antibiotics in farm animals can lead to the development of resistance in bacteria associated with the animal or with people who eat the animal. Such resistance can then be passed on to human pathogens by mechanisms of HGT.
Of major concern is the use of antibiotics as feed additives given to farm animals to promote animal growth and to prevent infections (rather than cure infections). The use of an antibiotic in this way contributes to the emergence of antibiotic-resistant pathogens and reduces the effectiveness of the antibiotic to combat human infections.
Antibiotic resistance in genetically modified crops
Antibiotic-resistance genes are used as "markers" in genetically modified crops. The genes are inserted into the plant in early stages of development to in order to detect specific genes of interest . e.g. herbicide-resistant genes or insecticidal toxin genes. The antibiotic-resistance genes have no further role to play, but they are not removed from the final product. This practice has met with criticism because of the potential that the antibiotic-resistance genes could be acquired by microbes in the environment. In some cases these marker genes confer resistance to front-line antibiotics such as the beta-lactams and aminoglycosides.
Inappropriate use of antibiotics in the medical environment
One problem is the casual use of antibiotics in medical situations where they are of no value. This is the fault of both health care workers and patients. Prescribers sometimes thoughtlessly prescribe 'informed' demanding patients with antibiotics. This leads to use of antibiotics in circumstances where they are of not needed, e.g. viral upper respiratory infections such as cold and flu, except when there is serious threat of secondary bacterial infection. Another problem is patient failure to adhere to regimens for prescribed antibiotics.
Patients and doctors need to realize their responsibility when they begin an antibiotic regimen to combat an infectious disease. There are several measures that should be considered.
� Patients should not take antibiotics for which there is no medical value (corollary: doctors should not prescribe antibiotics for which there is no medical value).
� Patients should adhere to appropriate prescribing guidelines and take antibiotics until they have finished.
� Patients should be give combinations of antibiotics, when necessary, to minimize the development of resistance to a single antibiotic (as in the case of TB).
� Patients need to be given another antibiotic or combination of antibiotics if the first is not working.
Combating antibiotic resistance
The following are recommendations to combat the development of antibiotic resistance in bacteria and other microorganisms.
Search for new antibiotics. To combat the occurrence of resistant bacteria, biotechnology and pharmaceutical companies must constantly research, develop and test new antimicrobials in order to maintain a pool of effective drugs on the market.
Stop the use of antibiotics as growth-promoting substances in farm animals. Of major concern is the use of antibiotics as feed additives given to farm animals to promote animal growth and to prevent infections rather than cure infections. The use of such antibiotics contributes to the emergence of antibiotic-resistant bacteria that threaten human health and decreases the effectiveness of the same antibiotics used to combat human infections.
Use the right antibiotic in an infectious situation as determined by antibiotic sensitivity testing, when possible.
Stop unnecessary antibiotic prescriptions. Unnecessary antibiotic prescriptions have been identified as causes for an enhanced rate of resistance development. Unnecessary prescriptions of antibiotics are made when antibiotics are prescribed for viral infections (antibiotics have no effect on viruses). This gives the opportunity for indigenous bacteria (normal flora) to acquire resistance that can be passed on to pathogens.
Finish antibiotic prescriptions. Unfinished antibiotic prescriptions may leave some bacteria alive or may expose them to sub-inhibitory concentrations of antibiotics for a prolonged period of time. Mycobacterium tuberculosis is a slow growing bacteria which infects the lung and causes tuberculosis. This disease kills more adults than any other infectious disease. Due to the slow growing nature of the infection, treatment programs last for months or even years. This has led to many cases on unfinished prescriptions and 5% of strains now observed are completely resistant to all known treatments and hence incurable.
Several other possible solutions have been proposed or implemented to combat antibiotic resistance.
In the pharmaceutical industry, past and current strategies to combat resistance have not been effective. Pharmaceutical companies are seeking new, less costly strategies to develop antibiotics.
A decrease in the number of prescriptions for antibiotics, especially in small children, is occurring.
Several countries such as the UK have regulations concerning the use of antibiotics in animal feed.Several other possible solutions have been proposed or implemented to combat antibiotic resistance.
In the pharmaceutical industry, past and current strategies to combat resistance have not been effective. Pharmaceutical companies are seeking new, less costly strategies to develop antibiotics.
A decrease in the number of prescriptions for antibiotics, especially in small children, is occurring.
Large scale public health education efforts are underway to stress the importance of finishing prescriptions. Indeed, in many places, failure to finish tuberculosis prescriptions can result in jail time.
Summary
The discovery of antibiotics was a leap in modern medicine. They have been able to stop the growth or kill many different kinds of microorganisms. However, bacteria have proven to be much more innovative and adaptive than we imagined and have developed resistance to antibiotics at an ever increasing pace. Bad practices and mismanagement have only exacerbated the situation. We could soon return to a state of medical health that was as dire as that which occurred prior to antibiotic use. However, with more research, education of the public, and well thought out regulations, the problems
can be solved. Several strategies are currently used to find new antibacterial compounds and new strategies are in development and trial.
Not only is there a problem in finding new antibiotics to fight old diseases (because resistant strains of bacteria have emerged), there is a parallel problem to find new antibiotics to fight new diseases. In the past three decades, many "new" bacterial diseases have been discovered (E. coli O157:H7 gastric ulcers, Lyme disease, toxic shock syndrome, "skin-eating" streptococci). Already broad patterns of resistance exist in these pathogens, and it seems likely that we will soon need new antibiotics to replace the handful that are effective now against these bacteria, especially as resistance begins to emerge among them in the selective environment antibiotic chemotherapy.
It is said that the discovery and use of antibiotics and immunization procedures against infectious disease are two developments in the field of microbiology that have contributed about twenty years to the average life span of humans in developed countries where these practices are employed. While the greater part of this span in time is probably due to vaccination, most of us are either still alive or have family members or friends who are still alive because an antibiotic conquered an infection that otherwise would have killed them. If we want to retain this medical luxury in our society we must be vigilant and proactive We must fully understand how and why antimicrobial agents work, and why they don't work, and realize that we must maintain a stride ahead of microbial pathogens that can only be contained by antibiotic chemotherapy.
END OF CHAPTER
No comments:
Post a Comment